Epidemiology and genotypic distribution of noroviruses in patients with acute gastroenteritis in developing and developed countries
Norovirus (NoV) is one of the major causes of viral sporadic and outbreak-associated gastroenteritis across all age groups both in developing and developed countries (1). With the implementation of rotavirus vaccines, the rotavirus-associated gastroenteritis cases have continuing declined and NoV infection has become the leading cause of acute gastroenteritis in children younger than 5 years of age (2-4). The NoV-associated mortality has been estimated in children aged less than 5 years in developing countries up to 218,000 deaths per year (5,6). In addition, NoV-associated deaths in adults in upper middle- and high-income countries have also been estimated at 2,000 to 13,000 deaths per year (7). Currently, no NoV vaccine is available, even though several NoV vaccine candidates are in the process of development (8).
NoV is currently divided into at least 7 genogroups based on phylogenetic analysis of the entire virus genome or of individual virus genes (9,10). Each genogroup is further subdivided into genotypes based on phylogenetic analysis of capsid (VP1) and polymerase (RdRp) gene sequences (11), and more than 40 genotypes have been identified (10). The genogroup I (GI), GII and GIV are found to infect human and cause gastroenteritis with the most prevalence of GII and GI, respectively (12). Globally, the prevalence of GII accounts for an average of 96.0% while GI with an average of 3.6%, and mixed infections of GI and GII with an average of 0.4% (12). Based on the capsid genotype, GII.4 is the most predominant genotype in most of the countries worldwide with the prevalence of 67.2% followed by GII.3, GII.6, GII.2, GII.7 at 16.3%, 3.8%, 1.2%, 0.6%, respectively, with small percentage of other GII genotypes (12). A systemic review and meta-analysis of NoV global prevalence in gastroenteritis cases indicated that the pooled prevalence of NoV in patients with acute gastroenteritis is 18% and the prevalence in low-mortality developing countries is more or less the same as those in developed countries (19% vs. 20%) (1). Additionally, the prevalences of NoV in these two settings are relatively higher than those in high-mortality developing countries (14%; P=0.058). Most recently, another systemic review and meta-analysis reported the NoV prevalence in gastroenteritis cases in developing countries at 17% and there are no significant differences in NoV prevalence by age groups or severity of symptoms (13).
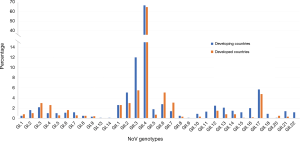
Regarding the frequency of NoV genotype distributions in developing (14-17) and developed (18,19) countries, generally, a great variety of NoV GI and GII genotypes are similarly distributed in the developing and developed countries except for some genotypes, GI.13, GI.14, GII.9, GII.11, GII.18, and GII.22 that are reported only from developing but not from developed countries (Figure 1). The GII.4 remains the most predominant genotype both in developing and developed countries with the average prevalence of 66.6% and 64.7%, respectively. The detection rates of GII.2, GII.3 and GII.17 are higher in developing than in developed countries, while those of GII.6 and GII.7 are higher in developed countries. Other GI and GII genotypes are also detected but at a small percentage.
Recently, Hossain and colleagues published an article in Clinical Infectious Diseases (20) describing the epidemiology and distribution of NoV genotypes in children admitted to a rural hospital and in healthy controls in Bangladesh from 2010 to 2012. Bangladesh is classified as one of the developing countries that NoV infection remains a public health problem. They reported the prevalence of NoV in cases with diarrhea at 18% and controls at 15% which is relatively lower than those reported previously by Nelson et al. (21), another group of investigators, who had also conducted a case-control study in Bangladesh at the same periods of time, 2010–2013. Nelson et al. reported the prevalence of NoV in cases with diarrhea at 19.6% and controls at 22.1%. The number of cases and controls of Nelson et al. study were about 2.7 times higher than those of Hossain et al. study. The frequency of detection of Nov GI and GII between these two studies are also different. In Hossain et al. study the GI and GII were detected at 15.0% and 85.0%, respectively, while those reported by Nelson et al. were 30.0% and 63.3%, respectively, and 6.7% were mixed GI/GII infections. Hossain et al. did not reported the mixed GI/GII infections in their study. Another point of the difference between the two studies is the difference in the number of different kinds of NoV genotypes circulating in pediatric patients in Bangladesh. Hossain et al. reported 7 GI and 12 GII genotypes compare to 6 GI and 18 GII genotypes reported by Nelson et al. It should be noted that seven genotypes of GII (GII.5, GII.8, GII.9, GII.12, GII.14, GII.22, GII.25) detected by Nelson et al. were not detected by Hossain et al. study. Conversely, one each of GI (GI.9) and GII (GII.16) genotypes detected by Hossain et al. study were not detected by Nelson et al. study. The discrepancy between these two studies is probably due to the differences in the sample size included in the studies, the sensitivity of the methods used for the detection and identification of GI and GII genotypes as well as the primer sets used by the two studies.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Ahmed SM, Hall AJ, Robinson AE, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 2014;14:725-30. [Crossref] [PubMed]
- Koo HL, Neill FH, Estes MK, et al. Noroviruses: the most common pediatric viral enteric pathogen at a large university hospital after introduction of rotavirus vaccination. J Pediatric Infect Dis Soc 2013;2:57-60. [Crossref] [PubMed]
- Doll MK, Gagneur A, Tapiéro B, et al. Temporal changes in pediatric gastroenteritis after rotavirus vaccination in Quebec. Pediatr Infect Dis J 2016;35:555-60. [Crossref] [PubMed]
- McAtee CL, Webman R, Gilman RH, et al. Burden of norovirus and rotavirus in children after rotavirus vaccine introduction, Cochabamba, Bolivia. Am J Trop Med Hyg 2016;94:212-7. [Crossref] [PubMed]
- Patel MM, Widdowson MA, Glass RI, et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 2008;14:1224-31. [Crossref] [PubMed]
- Lanata CF, Fischer-Walker CL, Olascoaga AC, et al. Global causes of diarrheal disease mortality in children <5 years of age: a systematic review. PLoS One 2013;8:e72788. [Crossref] [PubMed]
- Lindsay L, Wolter J, De Coster I, et al. A decade of norovirus disease risk among older adults in upper-middle and high income countries: a systematic review. BMC Infect Dis 2015;15:425. [Crossref] [PubMed]
- Cortes-Penfield NW, Ramani S, Estes MK, et al. Prospects and challenges in the development of a norovirus vaccine. Clin Ther 2017;39:1537-49. [Crossref] [PubMed]
- Tse H, Chan WM, Lam CS, et al. Complete genome sequences of novel rat noroviruses in Hong Kong. J Virol 2012;86:12435-6. [Crossref] [PubMed]
- Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015;53:373-81. [Crossref] [PubMed]
- Kroneman A, Vega E, Vennema H, et al. Proposal for a unified norovirus nomenclature and genotyping. Arch Virol 2013;158:2059-68. [Crossref] [PubMed]
- Hoa Tran TN, Trainor E, Nakagomi T, et al. Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants. J Clin Virol 2013;56:185-93. [Crossref] [PubMed]
- Nguyen GT, Phan K, Teng I, et al. A systematic review and meta-analysis of the prevalence of norovirus in cases of gastroenteritis in developing countries. Medicine (Baltimore) 2017;96:e8139. [Crossref] [PubMed]
- Kumthip K, Khamrin P, Maneekarn N. Molecular epidemiology and genotype distributions of noroviruses and sapoviruses in Thailand 2000-2016: A review. J Med Virol 2018;90:617-24. [Crossref] [PubMed]
- Mans J, Armah GE, Steele AD, et al. Norovirus Epidemiology in Africa: A Review. PLoS One 2016;11:e0146280. [Crossref] [PubMed]
- Zhou HL, Zhen SS, Wang JX, et al. Burden of acute gastroenteritis caused by norovirus in China: A systematic review. J Infect 2017;75:216-24. [Crossref] [PubMed]
- Supadej K, Khamrin P, Kumthip K, et al. Distribution of norovirus and sapovirus genotypes with emergence of NoV GII.P16/GII.2 recombinant strains in Chiang Mai, Thailand. J Med Virol 2019;91:215-24. [Crossref] [PubMed]
- van Beek J, de Graaf M, Al-Hello H, et al. Molecular surveillance of norovirus, 2005-16: an epidemiological analysis of data collected from the NoroNet network. Lancet Infect Dis 2018;18:545-53. [Crossref] [PubMed]
- Cannon JL, Barclay L, Collins NR, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 2017;55:2208-21. [Crossref] [PubMed]
- Hossain ME, Rahman R. Epidemiologic and genotypic distribution of noroviruses among children with acute diarrhea and healthy controls in a low-income rural setting. Clin Infect Dis 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Nelson MI, Mahfuz M, Chhabra P, et al. Genetic diversity of noroviruses circulating in a pediatric cohort in Bangladesh. J Infect Dis 2018;218:1937-42. [Crossref] [PubMed]
Cite this article as: Kumthip K, Khamrin P, Maneekarn N. Epidemiology and genotypic distribution of noroviruses in patients with acute gastroenteritis in developing and developed countries. Ann Res Hosp 2019;3:1.